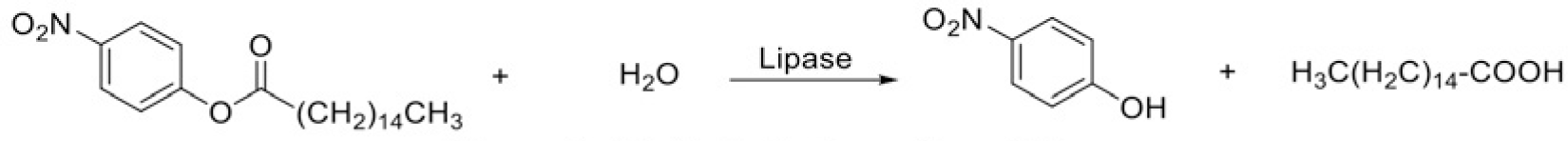1. INTRODUCTION
The obesity epidemic has become a burning issue in many healthcare systems in the world due to its dramatically increasing rate and severe impacts on the humans. According to WHO, there were 600 thousands of people had obesity in 2014, the number of obesity people is projected to reach more than 3 millions by the year 2030 [1]. Obesity increases two folds of the risk of obesity-related death between overweight individual and their normal weight counterpart. The most common diseases ralate to obesity are cardiovascular problems [2]. Moreover, 300.000 people die each year because of obesity complication [1, 3].
Obesity is the result of energy imbalance between calories consumed and calories expended. A present approach to treat obesity is inhibiting fat absorption, therefore reducing energy intake. Since fat is absorbed through the activity of pancreatic lipase, inhibiting this enzyme can be the strategy for development of anti-obesity agents [4, 5]. An available drug for the treatment of obesity acting like this way is Orlistat which originates from Streptomyces toxytricini [6, 7]. Although Orlistat is effective and popular in treating overweight, it has some noticeable side effects such as fatty stools, inability to control bowel movements [8]. Hence, it is necessary to find a drug that not only is more potent, but also has fewer side effects. Chalcone derivatives have risen as promising compounds which were demonstrated to be safe for people and easy to synthesize in the laboratory [9].
Pharmaceutical chemistry department in the University of Medicine and Pharmacy at Ho Chi Minh City, Vietnam has synthesized many chalcone derivatives and assessed these compounds in prostaglandin E2 inhibitory activity [10], antioxidant, anti-fungal activity [11, 12], antibacterial activity [10].
Human pancreatic lipase (HPL), which is synthesized and secreted into the duodenum through the system of the pancreas, is responsible for catalyzing the digestion of dietary triglycerides. From 50% to 70% of total dietary fats are hydrolyzed by this enzyme [5]. The main products are free fatty acids monoacylglycerols, fat soluble vitamins, bile salts, lysophosphatidic acid and diacylglycerols binding with cholesterol which are absorbed by enterocytes [13].
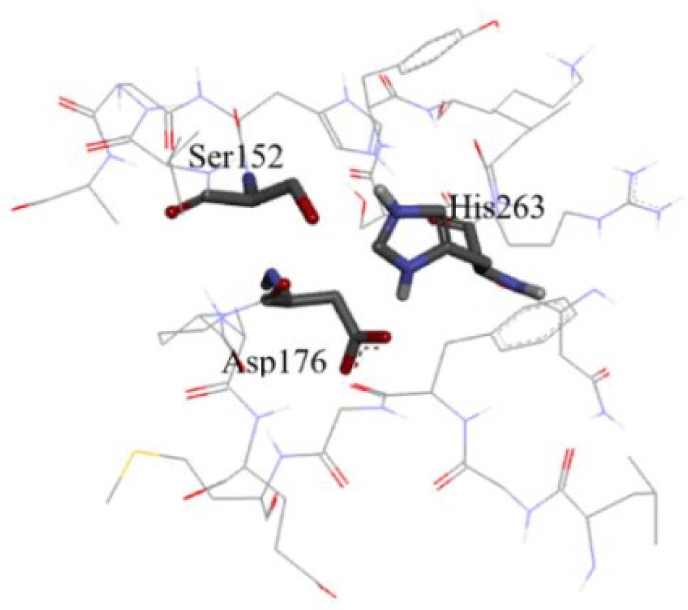
The structure of human pancreatic lipase was first isolated in 1974. It is a glycoprotein with the molecular mass of 46000 Dalton [14]. The primary structure of the HPL was built by analysis of cDNA clones isolated from a human pancreas cDNA library and be described as a single chain glycoprotein of 449 amino acids. The three-dimensional structure of HPL was determined by X-ray crystallography with the resolution of 2.46 A (PDB id: 1LPB) [15]. The encoded protein illustrated the similar identitiy of HPL with porcine and canine pancreatic lipase around 86% and 68% respectively. The polypeptide chain comprises two folding units, the larger N-terminal domain, constituting residues 1-336 and a C terminal domain containing residues 337-449, typical of b-sandwich type [16]. In the N-terminal domain, the binding site of HPL is described like a funnel with the size of 13 x 4.5 A at its base. The funnel consists of two hand walls with the left-hand wall being the lowest of all lipase binding sites from other species such as Candida Antarctica and Pseudomonas. Moreover, HPL is the only lipase whose left-hand wall is lower than both the left and right-hand walls in the front view. In addition, the amphiphilic component of Cys237 to Cys261, which is called a lid, lying in the left-hand wall. In the opposite side of the funnel, the P 5-loop constitutes the right-hand wall [17]. Inside the binding site, a triad which comprises His263, Asp176 and Ser152 acting as lypolytic site, (Figure 1). Among these residues, Ser152 plays the most essential role in the catalytic activity as the enzymatic activity will diminish if Serl52 is modified [17, 18].
With the aim of finding the potential compounds which have good inhibitory activity to pancreatic lipase, in silico approach was used first to orientate the strategy to synthesize chalcone derivatives. Biological testing was then conducted to evaluate the pancreatic inhibitory activities. The advanced point of this research is using the benzyl amino chalcone derivatives in the screening list, that have never been published for their pancreatic lipase inhibitory activity.
2. MATERIALS AND METHOD
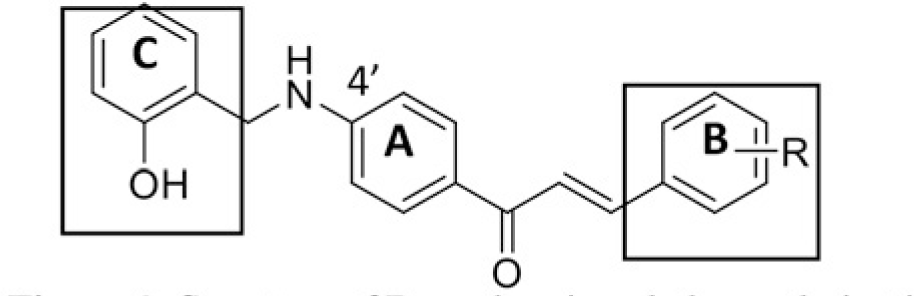
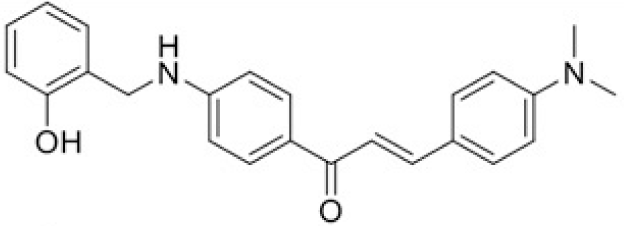
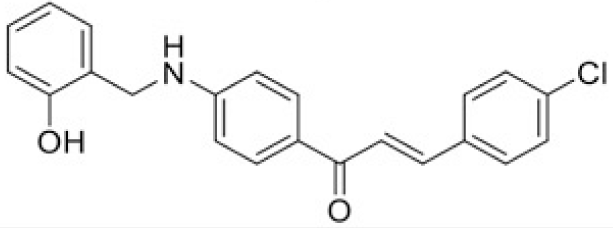
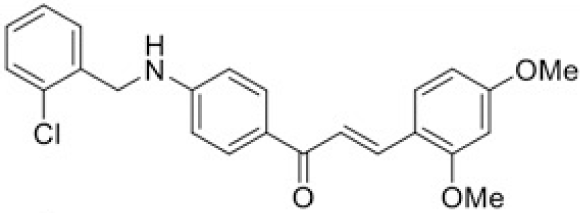
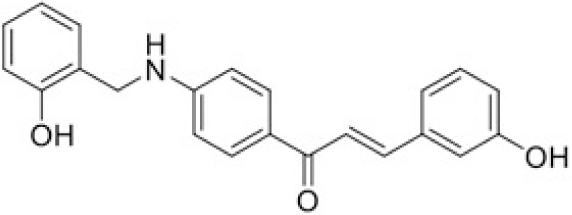
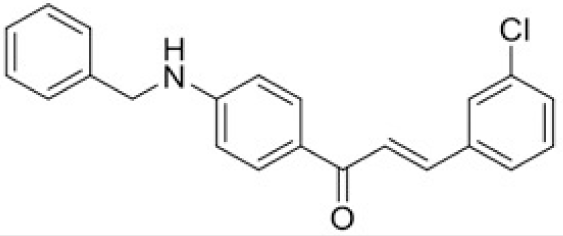
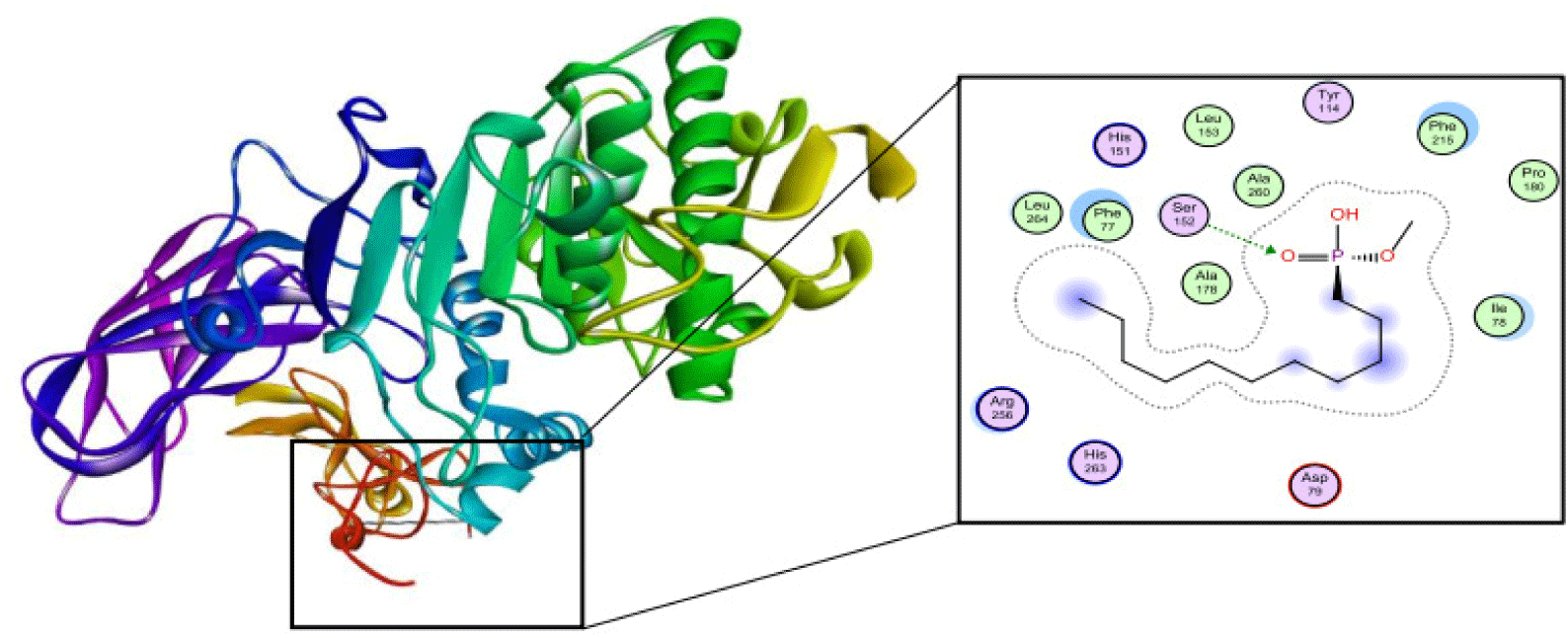
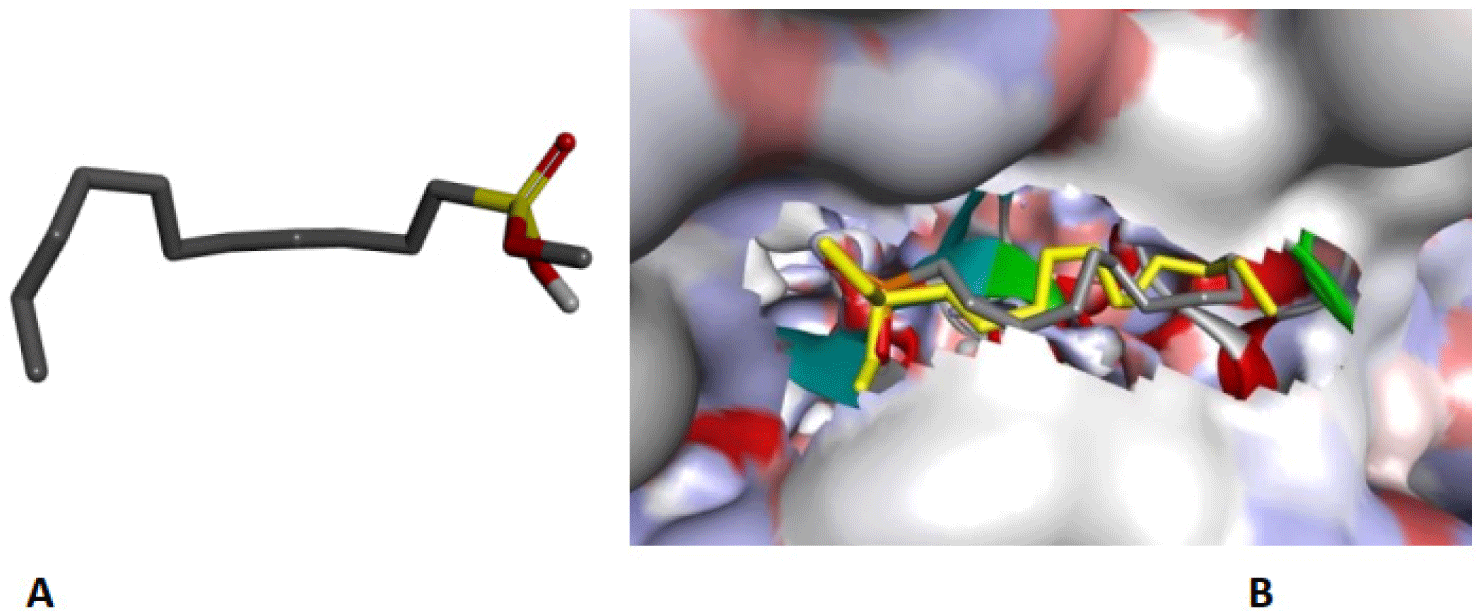
The structure of human pancreatic lipase (PDB id: 1LPB) was downloaded from Protein Data Bank as a therapeutic target [15]. The co-crystallized structure contains enzyme and methoxy undecyl phosphonic (MUP) acid as the inhibitor in the active site of the enzyme which contains the lipolytic triad (Figure 2). Water and ions were removed from the protein and protein was then protonated. It is hypothesize that the good inhibitory of MUP and Orlistat is attributable to the hydrophobic part of these two compounds. Therefore, a benzyl group is attached to chalcone to mimic the hydrophobic effect. Benzyl amino chalcone derivatives for screening in this study contain a chalcone core structure and a (2’-hydroxy) benzyl amino chain in 4’ position on ring A (Figure 3). Ring B is a heterocyclic ring or a phenyl ring with different substitution groups. Combining these features with available materials and facilities, 102 chalcone derivatives were generated and drawn by ChemBioDraw Ultra 13.0 [20] and converted to the 3D structure using OpenBabel software version 2.3.2 [21].
Co-crystallized ligand, chalcone compounds and Orlistat were docked into the target protein using Autodock Vina version 1.5.6 [22]. The grid box which had the size of 90 x 70 x 100 Å to cover both the binding site and MUP. The “exhaustiveness” and maximum energy difference were set as default in Autodock Vina (8 and 3 respectively). The results were evaluated by binding affinities and interactions of protein and ligands as well as the value of root-mean-square deviation (RMSD) between co-crystallized and docking ligands, which are likely to reproduce the experimental data and assess the precision of ligand preparation procedures and docking method. Binding affinity, interaction between ligands and target protein were used to rank the inhibitory activity of chalcone derivatives. The ligand which has the high binding affinity and strong interaction with protein is expected to show the good effect in inhibiting lipase enzyme.
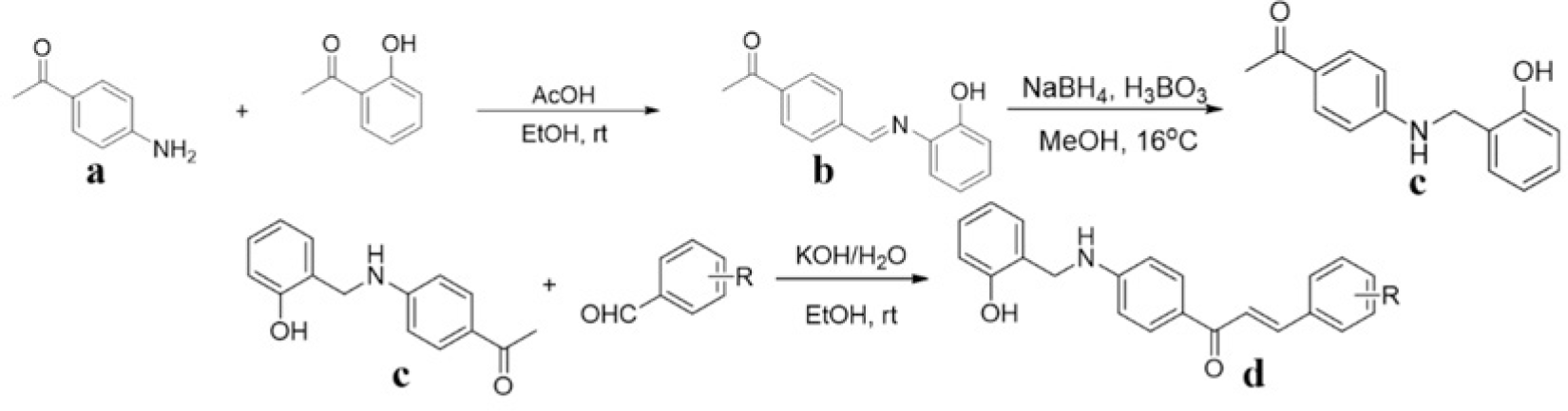
Synthesis process consisted of three reactions (Figure 4). Firstly, 4’-aminoacetophenone (a) reacted with salicylaldehyde to form (E)-1-(4-((2-hydroxy benzylidene)amino)phenyl) ethanone (b), then it was reduced by NaBH4 reagent to 1-(4-((2-hydroxy benzyl) amino)phenyl) ethanone (c) [23]. Finally (c) reacted with different benzaldehyde derivatives to form benzyl amino chalcone derivatives (d) [24].
The purity of the products were evaluated by physical properties (color, solubility, melting point), and thin layer chromatography. The structure of compound was determined by mass spectrometry, ultraviolet-visible spectroscopy and nuclear magnetic resonance methods. All results can be found in supplementary data.
Porcine pancreatic lipase is often used for pancreatic lipase inhibitory activity testing, as it contains 85% of homologous genes between human and porcine [6, 25]. Based on the absorbance variation of the reaction, p-nitro phenyl palmitate was used as a substrate for the lipase inhibition test [26]. The substrate, lipase and the inhibitor were mixed and the absorbance of forming p-nitrophenol (Figure 5) at wavelength of 405 nm was monitored and recorded. Repeat the same process with blank samples.
Percentages of inhibition were calculated by following formula in which ΔABlank is the absorbance of blank sample and ΔAtest is the absorbance of the test sample.
3. RESULTS AND DISCUSSION
The co-crystallized ligand of pancreatic lipase enzyme, MUP, was used as control compound for docking to determine the liability of docking procedure and the ability to reproduce the experimental data. MUP was redocked and the RMSD which measures the accuracy of docking model shows the value of 1.4976 A. This result indicates that the differences between the experimental and docking pose of MUP can be accepted (Figure 6). The binding site includes Ser152, Phe215, Arg256, His263 and Leu264.
102 chalcones were then docked into pancreatic lipase protein, and the top-binding affinity ligands are displayed in the Table 1.
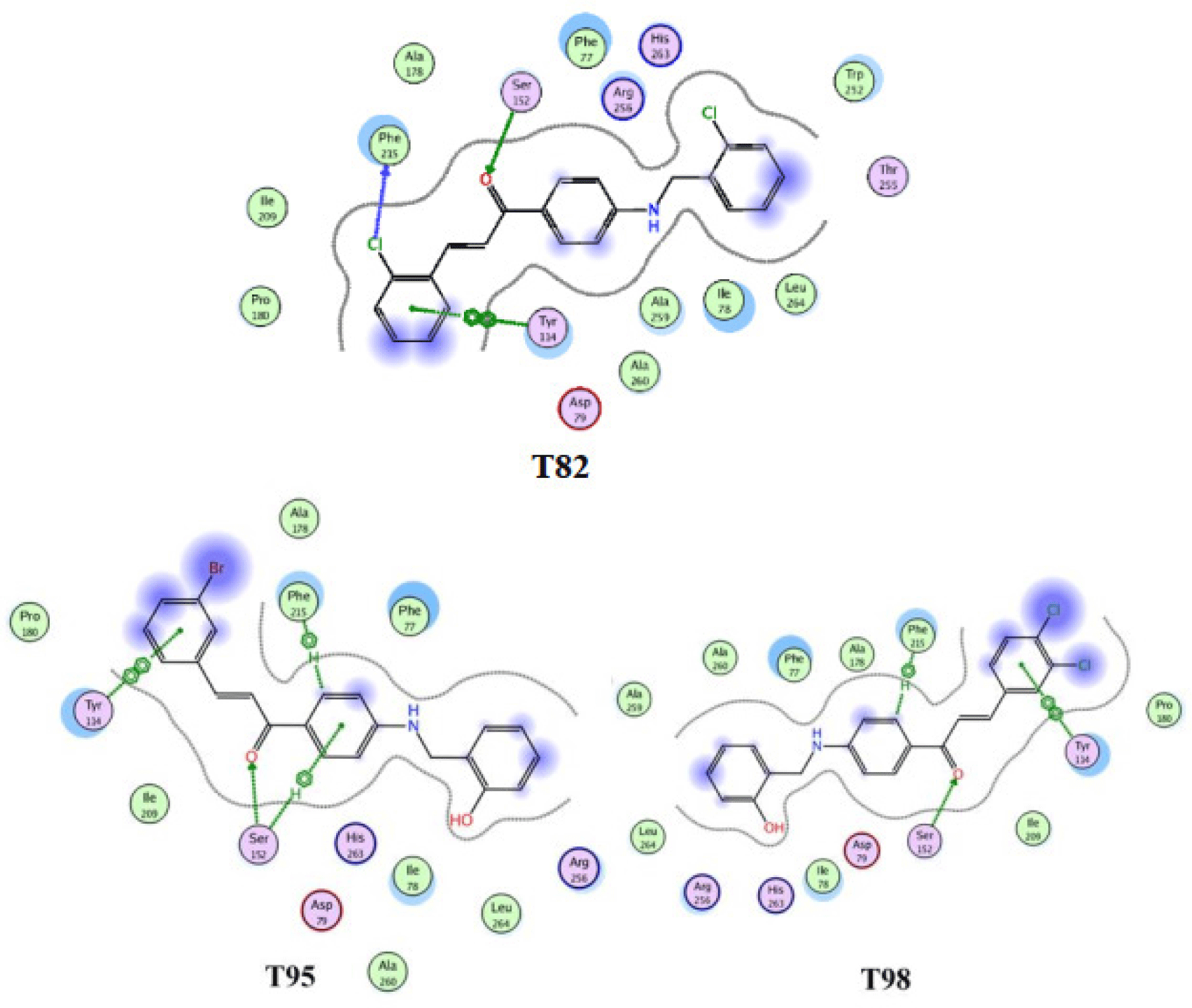
The docking results agreed with the finding of other researches that Seri 52 plays a significant role in hydrolyzing ester’s links of lipase due to its capability of forming hydrogen bonds with the carbonyl group of all chalcone derivatives. All ten ligands span along almost the whole length of the catalytic space in pancreatic lipase. Moreover, ring B of T98, T95, T82 also form n-n interaction with Tyr114 (Figure 7), which involve aromatic-aromatic interaction between two benzene rings of these chalcones and Tyr114. In addition, there is a hydrogen bonding between the chlorine atom of the ring B of the compound T82 with the residue Phe215. Furthermore, the hydrophobic regions (purple areas) are thought to stabilize the ligandprotein complex and contribute to the significantly good binding affinity of these three compounds.
Based on the binding affinity results, available materials and facilities, 14 structures were synthesized and their inhibitory activities were evaluated. The IC50 and docking scores of top 14 structures are illustrated in Table 2. Orlistat was used as control compound since it is the only FDA approved drug acting on lipase enzyme.
All 14 chalcone derivatives were successfully docked into the binding site of lipase enzyme (PDB id: 1LPB) which are shown in Table 2. The binding affinities of the chalcone derivatives varied from -8.6 (T76) to -10.1 kcal/ mol (T95 and T96)
Pancreatic lipase inhibitory activity tests showed that the IC50 values of chalcone derivatives were clearly higher compared to the value of Orlistat (2.8 μM). In detail, four derivatives (T42, T78, T101, T102) had the values of IC50 > 120 μM; eight other derivatives’ IC50 (T76, T96, T97, T48, T99, T45, T100, T43, T98) ranged from 60 μM to 120 μM. T97 and T95 ended the list with the lowest IC50 values of 50.51 and 57.89 μM, respectively.
The values of IC50 were converted into pIC50, then the correlation between docking scores and pIC50 was established using linear regression (Figure 8). R-Squared, which stands for the goodness of fit, shows the value of 0.6361.
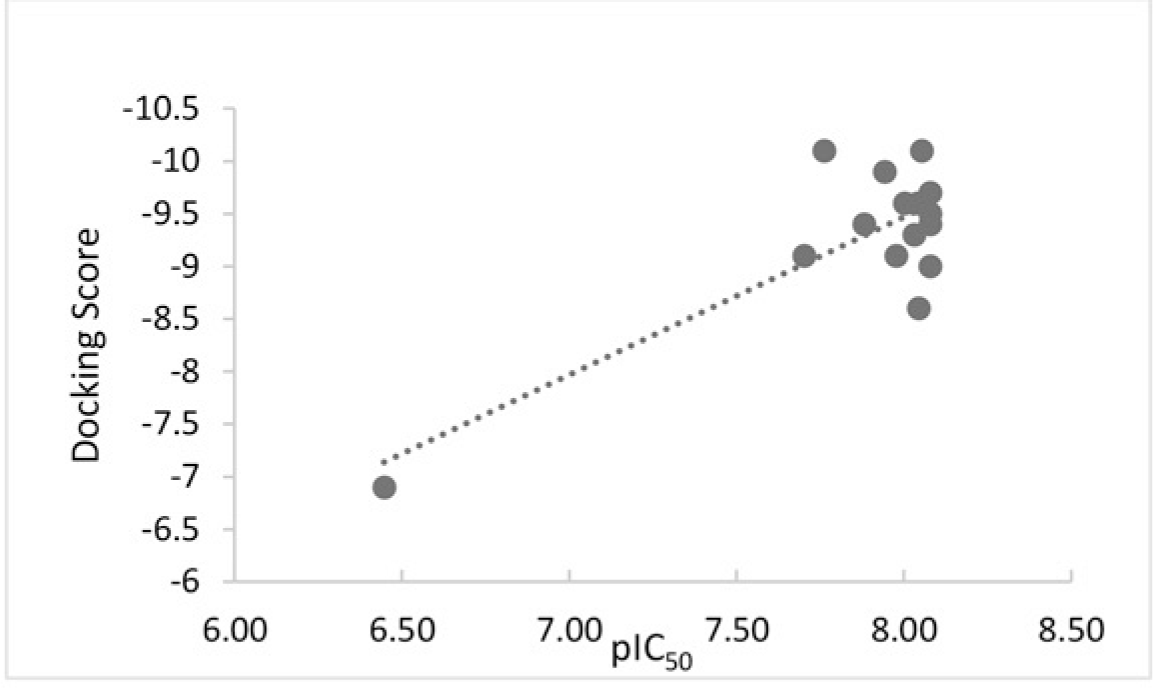
As can be seen from the the Figure 8, there is a moderate correlation (R2 = 0.6361) between the docking scores and IC50 of chalcone derivatives. The more negative the docking score is, the more chance of lipase inhibitory activity of the compound is stronger, which suggests the likelihood of using virtual screening as a practical method to orientate systhetic strategy and discovery new leads. Few exceptions in this correlation are also observed.
Although T97 structures did not have a good binding affinity (-9.1 kcal/mol), it showed a good effect in inhibiting lipase. In contrast, T98, T42, T78 derivatives whose binding affinities were among the most negative values (ranging from -9.5 to -10.1 kcal/mol), showed poor lipase inhibitory activities. In addition, Orlistat was in the same pattern which has less negative binding affinity but it shows a strong effect in inhibiting lipase (IC50 = 2.8 μM).
Based on the chemical structures and the values of IC50, the structure-activity relationships of chalcones were observed (Table 3). The fluoro group showed the good inhibitory effect when attached to compound T43 to form compound T97 which had the lowest IC50 value (50.51 μM). The change of the nitro group from para to meta position in the compound T45 to form T99 resulted in increasing of IC50 from 76.18 to 109.98 μM. Moreover, the replacement of methyl group in T101 by ethyl group to form T100 showed the decrease of IC50 values from > 120 μM to 100.64 μM.
| Structure |
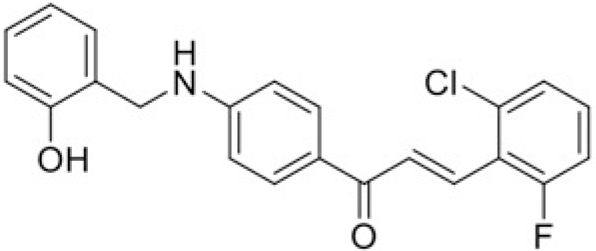
|
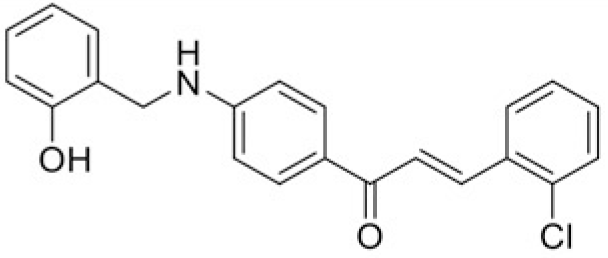
|
| Compound | T97 | T43 |
| IC50 (μM) | 50.51 | 87.7 |
| Structure |
|
|
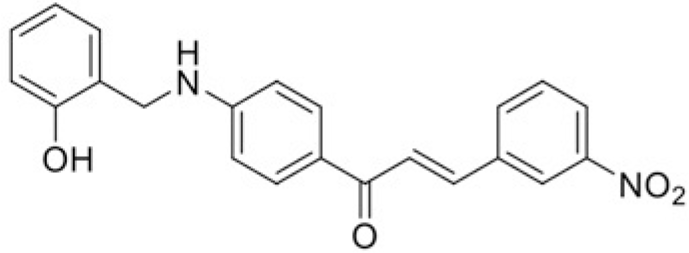
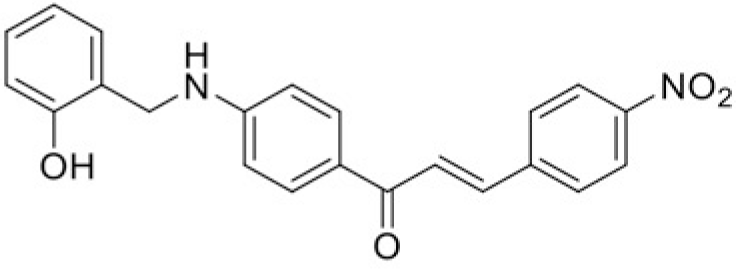
| Compound | T99 | T45 |
| IC50 (μM) | 76.18 | 109.98 |
| Structure |
|
|
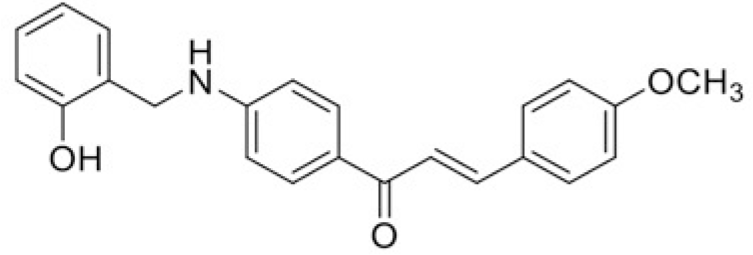
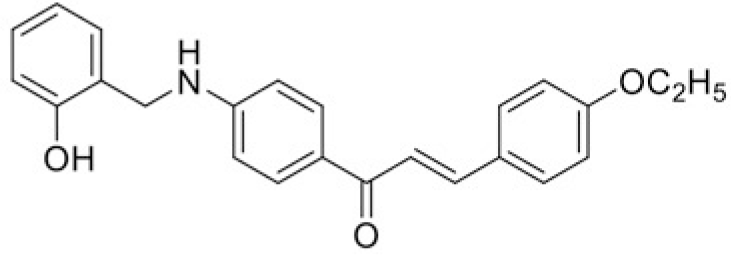
| Compound | T101 | T100 |
| IC50 (μM) | >120 | 100.64 |
The results showed that benzyl amino chalcone derivatives had moderate to weak pancreatic inhibitory activities compare to Orlistat and other flavonoids such as Theaflavin and its gallate derives whose IC50 ranged from 0.106 μM to 1.203 μM [27]. Other plant; extracts also showed strong activity against HPL. A research carried out by Sergent T and his colleagues found out that the aqueous ethanol extract from Capparis sicula Capparaceae, a Mediterranean dietary plant, showed strong inhibitory activity with an IC50 value of 0.53 ± 0.03 mg/mL. They also confirmed that the predominance in quantity of (±) catechin and rutin was contributed to the inhibitory activity [27, 28]. The strongly lipase inhibitory effect may be attributable to the synergetic effect of different chalcones in the extract. Moreover, it was possible to enhance the inhibitory activity of flavone by attaching a group of C-glucoside. The research group also confirmed that C-glycosyl flavones with two sugar moieties at C6 showed the highest inhibitory effect [7]. In addition, the existence of galloyl moieties within the structure of flavan-3-ols is responsible for the increase of HPL inhibition [29].
4. CONCLUSION
In this study, we screened 102 chalcone derivatives using in silico approach, synthesized 14 biologically potential derivatives and evaluated for their lipase inhibitory activities. The results showed that benzyl amino chalcone derivatives had moderate to weak pancreatic inhibitory activity. In addition, the molecular docking confirmed the key role of Ser152 in interacting with the ligands and the binding site consisting of two parallel hydrophobic walls and residues for potential n-n interactions. Hence, the hydrophobic moiety would play a critical role in forming a stable complex. A suitable benzyl amino chalcone derivatives should have those structural features for example a longer alkyl chain or more aromatic rings to become a lead compound for the anti-obesity drug discovery. Moreover, study also observed some structure-activity relationships which can employ to enhance the lipase inhibitory activity.